Key Resources
Merow et al. (2013) provides a very thorough introduction to Maxent modeling, and especially to what the various settings mean and how to set them.
RSpatial is a (nearly complete) set of lessons covering spatial data analysis in R, and including good tutorials for Maxent and other SDM approaches.
Best Practices in Species Distribution Modeling is another set of online notes, including R code.
From the same author (Adam B. Smith), a database of biodiversity databases.
Another workshop, from Lee-Yaw, is available on github. It’s a little old now, and doesn’t appear to be as comprehensive as Smith’s workshop above (maybe?). However, some people may prefer the more condensed format. Both authors have demonstrated expertise in the subject, so I don’t doubt that these are both reliable sources.
Technical Summaries
Phillips et al. (2017) provides a short discussion of recent developments of the Maxent application, and related statisical methods. See papers cited there for (even more) detailed information.
Elith et al. (2011) provides a statistical interpretation of the Maxent process; possibly superceded by Phillips et al. (2017) and the work cited therein, which was done post-2010.
Valavi et al. (2022) reproduce and extend the seminal paper of Elith (2006), showing that 16 years later Maxent remains one of the top performing SDM approaches (although some newer entrants may be slightly better now).
Sampling Bias
Sampling bias in occurrence data is an issue because it means we can’t be sure a species is detected under certain conditions because that’s its preferred habitat, or because those are the conditions in the locations we prefer to search.
The uniform sampling assumption does not require a uniformly random sample from geographic space, but instead that environmental conditions are sampled in proportion to their availability, regardless of their spatial pattern (Merow et al., 2013)
This problem can be addressed by thinning records (also called spatial filtering, Radosavljevic and Anderson, 2014), such that multiple records from within the same area are represented by only one or a few of the total records. This is a bit crude, but should remove the worst biases, such as a particular field station getting preferentially sampled by recurring visits from scientists or students, or general biases towards sampling roadsides and popular trails.
Lee‐Yaw et al. (2018) developed their own method to thin species records, using
kernel smoothing estimates to reduce the number of samples from a
neighbourhood, and selecting which samples to keep by identifying which
occupy novel environments. I don’t think this is widespread, and feels a
bit like overkill. 2025-05-05: This still isn’t widespread, but there is
evidence to support the importance of environmental filtering in model
accuracy (Varela et al., 2014). I need to investigate this further!
Another interesting approach in Atwater et al. (2018), who used a large dataset of gbif records to establish geographic (and presumably also environmental) bias in the full set of records, and used that to correct bias for individual species. Simply put, occurrence records for each species are weighted by the proportion of records for the entire set are found in that location (either geographic or a cell in a climate grid).
Subsampling based on raster grids is a simpler, more intuitive approach provided by Hijmans et al. (2017). It doesn’t account for the possibility that local density may be an accurate reflection of the niche requirements of a species, as the approach of Lee‐Yaw et al. (2018) does.
NB: See my extended discussion of thinning records on a grid
Grid-sampling:
library(terra)
## `occs` is a spatVector object containing the occurrence
## records as points
## wclim is a spatRaster object containing the environmental
## variables
## select one occurrence record per cell in the
## environmental raster layers::
lsOccs <- spatSample(occs, size = 1, strata = wclim)Aiello‐Lammens et al. (2015) provides an alternative approach based on imposing a minimum permissible nearest-neighbour distance, and then finding the set that retains the most samples through repeated random samples.
Thinning by Nearest-Neighbour:
## thin.par sets minimum distance in km
trichthin <- thin(data.frame(LAT = coordinates(trich)[, "Y"],
LONG = coordinates(trich)[, "X"],
SPEC = rep("tplan", nrow(trich))),
thin.par = 2, reps = 1, write.files = FALSE,
locs.thinned.list.return = TRUE) Radosavljevic and Anderson (2014) show that unfiltered/unthinned data produces elevated assessment of model performance, as a consequence of over-fitting to spatially auto-correlated data. So filtering works.
See also Boria et al. 2014 (unread), Varela et al. (2014)
Merow et al. (2013) provide two more rigorous approaches, depending on whether or not data on search effort is available. When search effort is known, it can be used to construct a biased prior.
When search effort is unknown, we can create a biased background sample to account for bias in presence data, via Target Group Sampling. Under TGS, records that are collected using the same surveys/methods as the focal species are form the background points. i.e., the set of all herbarium records in GBIF may be an appropriately biased background for any one of those plant species. This assumes that the target plant is collected/detected at the same rate as the reference set. It may be appropriate to subset the reference set to increase the likelihood of this being true: use only graminoids as biased background for sedges, or woody plants as background for a tree?
Spatial Bias
Elith et al. (2011) – The area of individual cells in raster layers projected in Lat-Lon coordinates are not equal. This can be corrected by:
- project the grids to an equal area projection
- create a ‘bias grid’ that can be used to weight background samples
- create a background sample with appropriate sampling weights
Study Extent
Discussed extensively in Barve et al. (2011). They identified three general approaches to consider:
- Biotic regions (ecozones etc). A good compromise between biological realism and tractability
- Niche-model reconstructions: back-project a niche model over the appropriate time period (i.e., previous glacial maximum or interglacial) to identify the area that the species could have occupied over an extended period. Nice idea, but a real risk of circularity?
- Detailed simulations. Sounds great, but I think if we had enough data to properly parameterize such a model, we wouldn’t need to resort to sdms in the first place.
If you wanted to improve on biotic regions, things to consider in developing a more rigorous approach should include:
- Dispersal characteristics of the species
- Crude estimate of the niche (again, circularity?)
- Establish relevant time span
- Identify relevant environmental changes
Soberón (2010) is often cited together with Barve et al. (2011), but the latter provides more explicit discussion of best practices for SDM model construction. I think the deference to Soberon is probably due to their creation of the BAM model (in earlier publications), which Barve’s system is based on (Biotic, Abiotic, Movement).
Merow et al. (2013) provide a shorter discussion, and emphasize matching the study extent to the biological question of interest. Prioritizing sites for protection within the range of a species should constrain the extent to the existing range of the species; evaluating invasion potential should use an extent large enough to encompass the areas of concern (i.e., global, or continental scale for novel invasives).
Background Points: Number and Bias Grids
The number of background points should be large enough to comprehensively sample (and hence represent) all environments in the region of interest. (Valavi et al., 2022)
If computational resources aren’t limiting, the ‘gold standard’ would be to use every cell in the background extent. When that’s not feasible (i.e., most of the time!), Valavi et al. (2022) recommend using 50,000 points. This is based on empirical tests on a large number of species which showed that model AUCROC approaches that of the ‘gold standard’ when 50K background points are used. This comes at a cost of doubling the computation time compared to sampling 10K background locations. They also note that choosing 50K, rather than 35K or 70K, is somewhat arbitrary.
This updates previous work that recommended sampling 10K background points (Phillips and Dudík, 2008; Barbet-Massin et al., 2012).
Variable Selection
Variables == predictors, the spatial layers used as the environmental/dependent variables in the model.
Interesting discussion in Guisan et al. (2017) (section 6.4, page 102+): variables that are measured most accurately often/usually are only indirectly related to a species’ niche; e.g., elevation, slope, aspect. Very precise and accurate spatial layers are available for these.
Variables with a direct relationship to a species niche are referred to as “proximal”. These include temperature, moisture, soil type etc, and are usually created through interpolation from sparse reference points (weather stations). This involves unavoidable error propagation and imprecision.
Over small extents, it may be preferable to use indirect variables, as they offer greater precision in quantifying the local environment. However, as extent increases, the relative value of direct variables increases. The indirect variables are likely not stationary on large scales - a species relationship to slope and elevation are likely different in southern US vs northern Canada, for instance. On the other hand, a species relationship to temperature, however coarsely it is mapped, is likely similar across its geographic range.
Elith et al. (2011) point out that Maxent’s built-in variable selection (via L1-regularization) is reliable, relatively insensitive to correlation among variables, and model performance may actually be degraded by imposing additional model selection procedures prior to running Maxent! They do suggest that sticking to proximal variables is preferable when projecting models to novel contexts.
Worldclim
One of the most common sources of climate data for SDMs is WorldClim. There are actually three distinct data sets available from Worldclim:
- Historical climate data
- compiled from a combination of weather stations and databases, and further refined with satellite imagery (Fick and Hijmans, 2017)
- climate normals (30 year averages for the period 1970-2000) including monthly minimum, maximum, average temperature; monthly total precipitation; 19 bioclim variables (biologically meaningful values calculated from monthly temperature and precipitation values).
- Historical monthly weather data (1950-2024)
- downscaled from CRU (Harris et al., 2020)
- 10-year averages for monthly minimum, maximum and average temperatures; monthly total precipitation.
- Future climate projections
- downscaled & bias corrected data from CMIP6, including a variety of global climate models (GCM) and shared socio-economic pathways (SSP).
- 20-year averages for monthly minimum, maximum and average temperatures; monthly total precipitation; bioclim variables.
Transferability
Petitpierre et al. (2017) explicitly tested different approaches to model selection for use in projecting models in space and time. They recommend modelers should use a small number of proximal variables, or, particularly when very few observations are available for training, the first few PCA axes of a larger set of environmental variables. PCA axes are orthogonal (i.e., not collinear) by construction, but interpretation may be tricky if they incorporate a large number of variables.
Since we rarely have much information on proximal variables (beyond understanding that temperature and moisture are generally important), Petitpierre et al. (2017) recommend a suite of “State of the Art” (SOA) variables that have been shown to be useful for plants. These include:
- BIO 01: annual mean temperature
- BIO 04: temperature seasonality
- BIO 11: mean temperature of coldest quarter
- BIO 10: mean temperature of warmest quarter
- BIO 15: precipitation seasonality
- BIO 16: precipitation of wettest quarter
- BIO 23: moisture index seasonality
- BIO 28: annual mean moisture index
Note the last two aren’t included in the WorldClim/Bioclim archives, and the numbering above BIO 19 isn’t necessarily consistent (at least between different sources). These variables are available from https://www.climond.org, but that appears to be an outdated source relative to Worldclim? Worldclim currently offers more and more recent climate projections, but only for the first 19 variables. The Climond data is currently is also only available at 10’ and 30’ resolution, while Worldclim provides data (without the moisture indices) down to 30" resolution.
At a global scale, annual mean temperature and temperature of the warmest quarter are (not suprisingly) highly correlated, and the moisture indices are moderately correlated with precip var and precip wet quarter (again, not suprising). So it might be reasonable to use a reduced set of the SOA variables in studies that aim to generate transferable models: BIO 01, BIO 11, BIO 15, BIO 16 (and maybe BIO 23).
Beware temperature/precipitation ratios!
Booth (2022) found some very interesting problems with climate variables that mix temperature and precipitation (e.g., temperature of wettest quarter, precipitation of warmest quarter). In some cases these can generate abrupt gradients that reflect numerical oddities of the ratios rather than any biologically meaningful pattern. If you use the SOA variables this isn’t a concern, but if you are using combination ratios, be sure to check that paper for details.
Filtering Collinear Variables
This section is out of date and needs to be updated and trimmed!
I will need to reconsider whether evaluating collinearity is something I should continue to do. In the meantime, I leave the following notes on how to do it:
Merow et al. (2013) identify two general approaches to selecting variables. The machine learning approach is based on the understanding that the Maxent algorithm will, by design, select the most useful variables and features, so we can include all reasonable variables.
However, this probably only applies when the objective is to provide accurate predictions of occurrences in the same context in which the model is built. Efforts to understand the environmental constraints on that distribution, or projecting it to a new context, will be potentially confounded when the model includes correlated variables.
To minimize this problem, Merow et al. (2013) recommends taking a statistical approach (i.e., treating a Maxent model as a ‘conventional’ statistical model). In this case, they recommend prescreening variables to limit colinearity, and emphasize biologically relevant variables. This should produce more parsimonious and interpretable models.
Pairwise correlations can be used to identify pairs or groups of variables
that are highly correlated. ENMTools (Warren et al., 2019) provides several
helper functions for this, including raster.cor.matrix,
raster.cor.plot.
I prefer using hclust based on 1 - abs(cor) to visualize correlated
groups:
## "predictors" is a raster stack
## Calculate correlations:
cors <- raster.cor.matrix(predictors) # from ENMTools
threshold = 0.7 ## the maximum permissible correlation
dists <- as.dist(1 - abs(cors))
clust <- hclust(dists, method = "single")
groups <- cutree(clust, h = 1 - threshold)
## Visualize groups:
plot(clust, hang = -1)
rect.hclust(clust, h = 1 - threshold)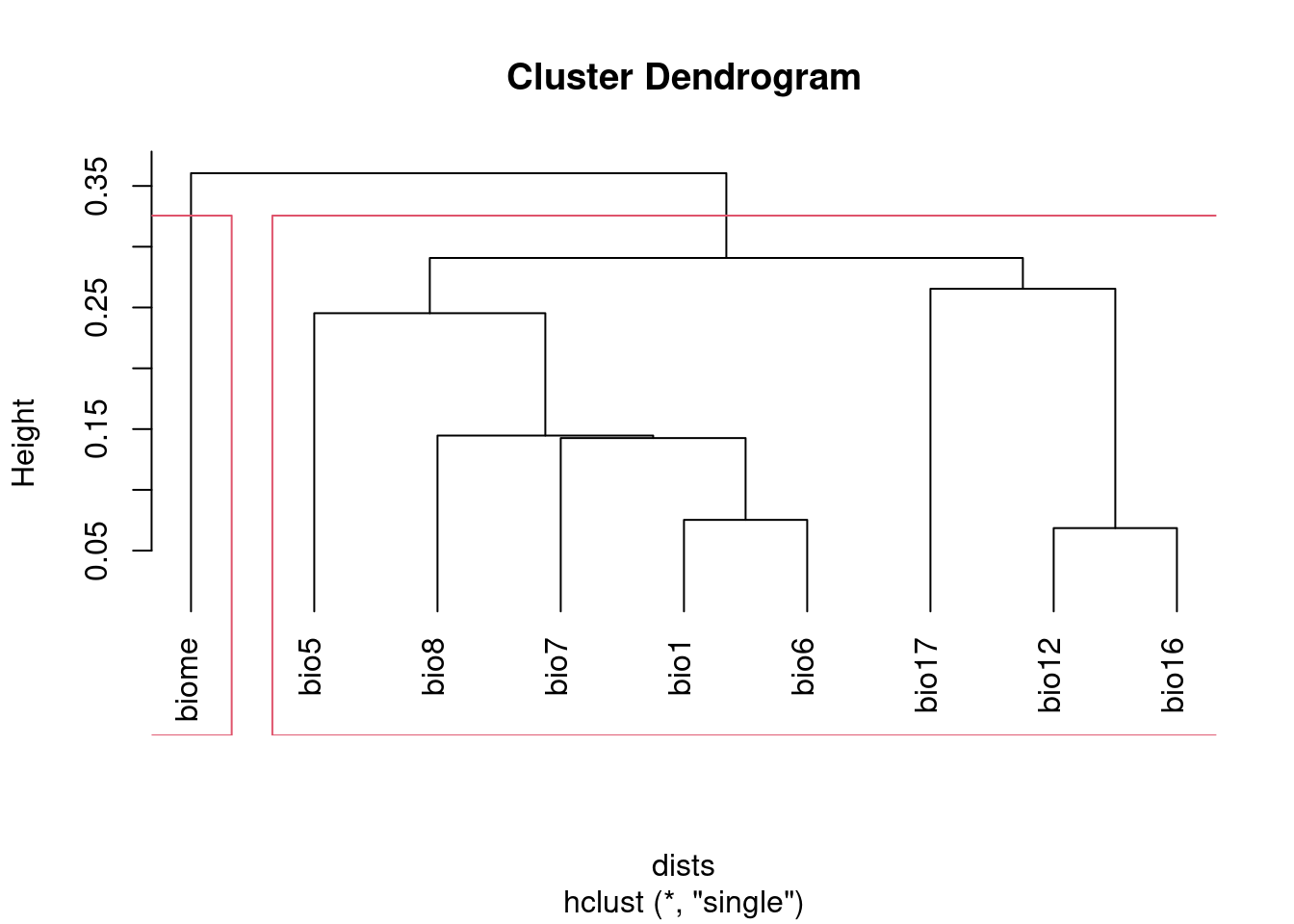
## Print the groups:
groups## bio1 bio12 bio16 bio17 bio5 bio6 bio7 bio8 biome
## 1 1 1 1 1 1 1 1 2NB I set the method argument to "single". This ensures that all the
variables with correlations above the threshold will be captured together
in a cluster. The default method, "complete", creates clusters that may
include variables with correlations above the specified threshold. See
discussion of hierarchical agglomerative clustering (section 8.5) in
Legendre and Legendre (2012) for details, if you want to know more.
After running this, groups will identify which cluster each variable
belongs to. Keep at most one variable from each group.
This doesn’t absolutely guarantee that variables in different groups will
have correlations less than threshold, but in most cases this will be
true. When it isn’t, the highest inter-group correlation will still be very
close to the threshold. If you’re concerned, double check the correlations
of the variables after you’ve picked them.
Alternatively, you can use cutree to select the number of groups you
want, then pick a variable from each group. Since this doesn’t use a
threshold, you’ll have to make sure the number of groups you pick is equal
to or lower than the number of groups generated using the threshold
approach.
## "predictors" is a raster stack
## Calculate correlations:
cors <- raster.cor.matrix(predictors) # from ENMTools
groupNum = 3 ## the desired number of groups
dists <- as.dist(1 - abs(cors))
clust <- hclust(dists, method = "single")
groups <- cutree(clust, k = groupNum)
## Visualize groups:
plot(clust, hang = -1)
rect.hclust(clust, k = groupNum)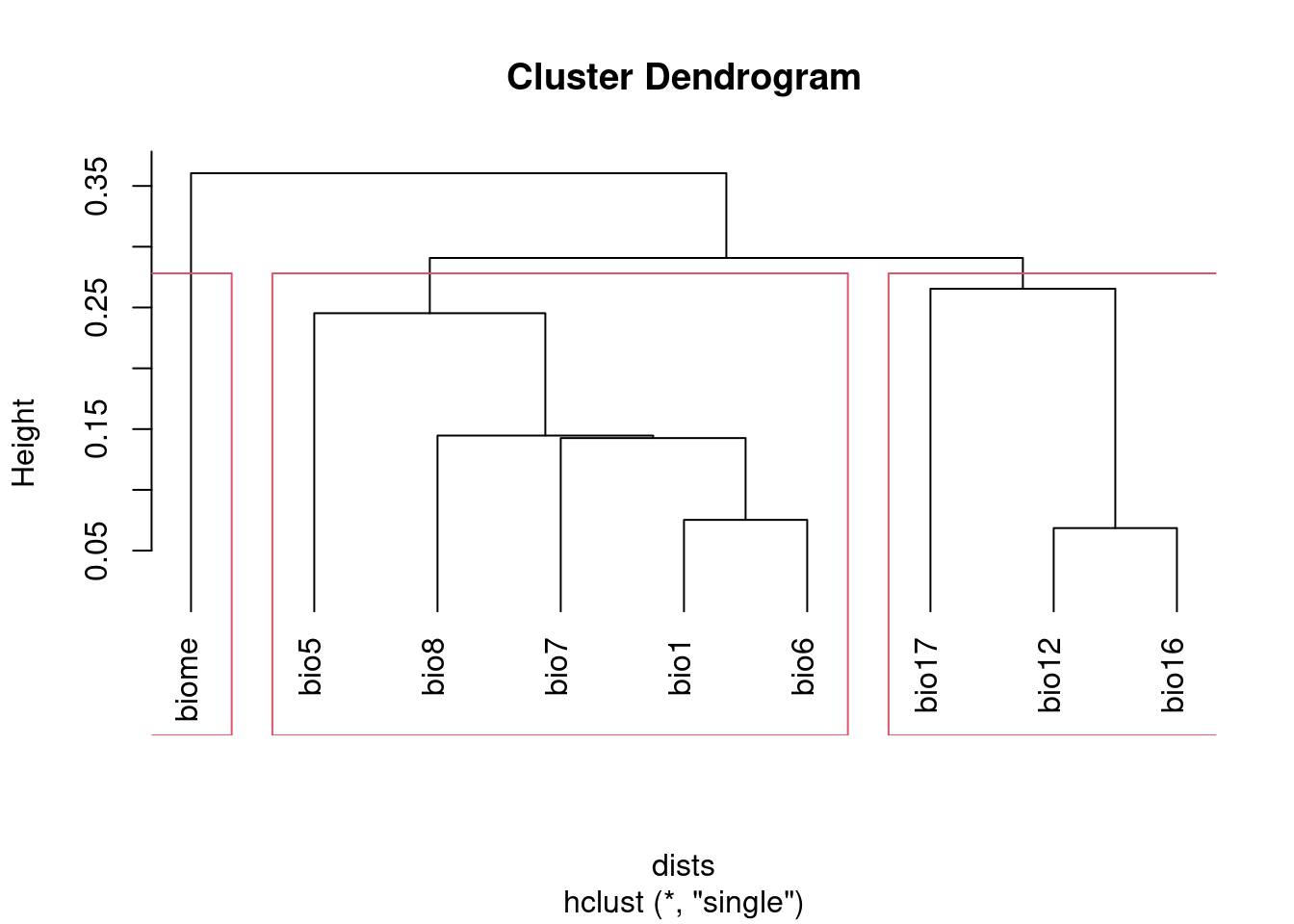
## Print the groups:
groups## bio1 bio12 bio16 bio17 bio5 bio6 bio7 bio8 biome
## 1 2 2 2 1 1 1 1 3However, neither of these approaches will address multicollinearity among
three or more variables. Guisan et al. (2017) suggest using the function
usdm::vif instead, which calculates variable inflation. They recommend
keeping the vif values under 10, but different authors will use cutoffs
from 5-20.
Feature Selection
Features == the statistical models used to fit the variables to the response variables (presences). i.e., linear, quadratic, product, hinge, threshold, categorical.
Note that hinge is essentially a superset of linear and threshold
features, so if you have hinges, the other two are redundant
(Elith et al., 2011). As of version 3.4.0, threshold featues are not included
by default; experience has shown that this improves model performance, and
produces simpler, more realistic models (Phillips et al., 2017). Similarly,
product features appear to contribute very little to model performance,
given the added complexity.
Merow et al. (2013) recommend selecting features on biological grounds. They provide a short discussion, noting that the fundamental niche is likely quadratic for most variables over a large enough extent, but may be better approximated by a linear function if the study extent is truncated with respect to the species’ tolerance for that variable (a la Whittaker). Interesting ideas, but not much to go on unless you actually do know a fair bit about your species.
Warren and Seifert (2011) describe a process for selecting features to keep/include in the model (linear, quadratic, polynomial, hinge, threshold, categorical). It uses the AIC to identify the optimal combination. Easy and quick to do with the ENMEval package (note that many references cite ENMTools for these tests, but they’ve been moved to ENMEval nowadays).
NB applying different spatial filtering/thinning to your data can produce different ‘optimal’ models (i.e., different retained features and regularization value), as determined by the AIC criterion.
NB The enmeval function only evaluates AIC for models with an
appropriate number of parameters. If you have a low number of observations,
a low regularization (i.e., permissive approach to including parameters),
and complex/high-parameter models (especially hinge features), the AIC
values will be reported as NA. These models are overfit, and as such you
shouldn’t use them with your data. Explanation on Maxent discussion
list.
Regularization
Regularization is used to penalize complexity. Low values will produce models with many predictors and features, with 0 leading to all features and variables being included. This can lead to problems with over-fitting and interpretation. Higher regularization values will lead to ‘smoother’, and hopefully more general and transferable models. There will be a trade-off between over- and under-fitting.
The default values in Maxent are based on empirical tests on a large number of species. These are probably not unreasonable, but it’s pretty standard to mention that they’re a compromise, and we improved them for our the needs of our particular species and context by doing X (for various values of X).
The approach of Warren and Seifert (2011) (see previous) can be used here as well, testing a range of regularization (aka beta) values, and selecting the one that generates the lowest AIC. It may also be worth selecting the simplest model that is within a certain similarity of the ‘best’ model? That’s more to explain to reviewers though.
Warren and Seifert’s simulations demonstrate that models with a similar number of parameters to the true model produce more accurate models, in terms of suitability, variable assessment, and ranking of habitat suitability, both for the training extent and for models projected in space/time. Furthermore, AIC and BIC are the most effective approaches to model tuning to achieve the correct number of parameters.
Radosavljevic and Anderson (2014) also consider the impact of the regularization parameter on over-fitting. They find that the default value often leads to over-fitting, especially when spatial auto-correlation is not accounted for in model fitting. They conclude that regularization should be set deliberately for a study, following the results of experiments exploring a range of potential values.
Note that specifying the regularization is done via the betamultiplier
argument, which applies to each of the different feature classes. That is,
the actual regularization value will be set by Maxent automatically for
each class, subject to the multiplier value specified by the user. We don’t
set the regularization values for each class directly (which is possible
via the options beta_lqp, beta_threshold etc. (Phillips, 2017),
although Radosavljevic and Anderson (2014) suggest experiments to explore this
should be done.
Output type
Raw: values are Relative Occurrence Rate (ROR) which will sum to 1
over the extent of the study. Merow et al. (2013) considers this to be a
reasonable interperetation of the Maxent output, but the actual values can
be difficult to interpret; they produce maps that “do not often match
ecologists’ intuition about the distribution of their species”
(Phillips et al., 2017).
Logistic: attempts to provide an accurate estimate of the probability that the species is present, given the environment. Monotonically related to raw values; site rank is identical for these measures. Based on the assumption that there is a 50% probability that a species will be present at a site with ‘average’ conditions for the species. This assumption is problematic and unrealistic according to Merow et al. (2013). However, Elith et al. (2011) prefer logistic output, and discuss justification for preferring it over raw values.
Cumulative: the sum of all cells with <= to the raw value of the cell. Rescaled to range from 0-100.
Complementary Log-Log (aka CLOGLOG): the standard output from version 3.4.0 onwards. Generally similar to the logistic output, but tending to give slightly higher values. There is a stronger theoretical justification for CLOGLOG than logistic, as summarized in Phillips et al. (2017). CLOGLOG provides an estimate of the probability of presence, but with the caveat that this probability is based on an arbitrary quadrat size (similar to the prevalence assumption made with logistic).
Merow et al. (2013) recommends sticking to Raw whenever possible, which means using the same species in the same extent. Note that the raw values will change for different extents, even for identical models, so they can’t be compared across projections without additional post-processing.
Cumulative is preferable when defining/describing range boundaries, or otherwise dealing with omission rates.
Elith et al. (2011) prefer to use the logistic, which they present as a biologically reasonable estimate. However, it will be a problem in comparisons among species with different prevalence on the landscape, as it assumes identical (and arbitrary) prevalence.
Following (Phillips et al., 2017), CLOGLOG now seems to be the most intuitive output to use in most cases.
Evaluation
AUC
AUC assesses the success of the model in correctly ranking a random background point and a random presence point; that is, it should predict the suitability of the presence point higher than the background point. It is threshold-independent.
Lobo et al. (2008) identified five problems with AUC:
- it ignores the predicted probability values and the goodness-of-fit of the model;
- it summarises the test performance over regions of the ROC space in which one would rarely operate;
- it weights omission and commission errors equally;
- it does not give information about the spatial distribution of model errors; and, most importantly,
- the total extent to which models are carried out highly influences the rate of well-predicted absences and the AUC scores.
Additionally, Radosavljevic and Anderson (2014) point out that AUC doesn’t assess over-fitting or goodness-of-fit; rather, it is a measure of discrimination capacity.
However, comparing the difference in AUC for the training and testing data does give an estimate of overfitting. If the model fit perfectly, without overfitting, the AUC should be identical. It won’t be, and the difference reflects the degree to which the model is over-fit on the training data. In other words, the extent to which the model is fit to noise in the data, or environmental bias, if geographic masking is used in the k-fold partitions.
Boyce
Boyce et al. (2002) proposed an index that compares the predicted and expected number of occupied sites with the suitability value of those sites:
sites are first sorted from lowest to highest suitability
then they are binned into groups of equal frequency
the number of actual occurrences for each bin are tabulated
the Spearman-rank correlation between bin rank and occurrence number is then calculated; if the model is good, we expect increasing numbers of occurrences for higher-ranked bins
Hirzel et al. (2006) evaluated a variety of SDM evaluation measures; on data sets with more than 50 presences, most evaluators had > 0.70 correlation with each other. Which is a little reassuring I suppose? They used AUC on presence/absence data as the ‘gold standard’, and found that the continuous Boyce index (which uses presence-only data) performed best.
This can be calculated with the function ecospat::boyce(), which takes a
raster of suitability values and a matrix or data.frame containing the
coordinates of presences. Note that as of 2025-05-06 there is a bug in
this function, and the
values it produces are note correct! Hopefully this will be fixed soon.
Related discussion in Phillips and Elith (2010), who note that the Boyce index is an example of their presence-only calibration plot (POC plot).
Thresholds
Radosavljevic and Anderson (2014) Threshold-dependent evaluation requires identifying a threshold in values predicted by the model to generate a binary suitable/unsuitable map. Setting the threshold to the lowest predicted value for a presence location may produce undesireable results if the lowest values is associated with an observation from an extreme outlier. More robust is setting the threshold to a particular quantile (10%), to exclude weirdos from establishing what’s suitable.
Again, if the model is perfectly fit, the omission rate in the testing data should be the same as in the training data. That is, setting the threshold at 10% to create the binary suitability map, we expect the omission rate in the test data to be 10%. Higher omission in the testing data reflects over-fitting (noise and/or bias).
For presence-only data commission error is unknown/unknowable. Accordingly, Radosavljevic and Anderson (2014) defined an optimal model as one that “(1) reduced omission rates to the lowest observed value (or near it) and minimized the difference between calibration and evaluation AUC [i.e., minimized over-fitting]; and (2) still led to maximal or near maximal observed values for the evaluation AUC (which assesses discriminatory ability). When more than one regularization multiplier fulfilled these criteria equally well, we chose the lowest one, to promote discriminatory ability (and hence, counter any tendency towards underfitting).”
Cross-validation
Radosavljevic and Anderson (2014) evaluated cross-validation using random k-fold partitions, geographical structuring, and geographic masking of partitions. Random partitions suffer from preserving biases in the training data in the testing data.
Geographic structuring, which uses occurrences from a pre-defined geographic area (rather than a random sample) as the test set, introduces additional spatial bias, and should be avoided. However, geographic structuring combined with masking (which excludes both presences and background from the specified geographic region from the test set) may substantially reduce overfitting, and yields more realistic models than random partitions.
Checkerboard partitions offer a nice compromise - this is geographic structuring and masking on a fine scale, and so should reduce spatial correlation between training and testing data. A version was used by Pearson et al. (2013), without a lot of discussion. Functions to do checkerboard cross validation are provided by Muscarella et al. (2014), but without a lot of discussion. The cited references suggest this might be intended more for species with limited occurrence data? Also, as implemented it looks like they only allow for 2-fold and 4-fold cross-validation. I’m not sure there’s any reason not to use checkerboards to do 9- or 16- fold cross validation?
Spatial Null Models
Not often used, but see Rodríguez-Rey et al. (2013), and discussion of Bahn and McGill (2007).
The dismo package (Hijmans et al., 2017) provides the function geoDist to
serve as a spatial null model. Provided a matrix of occurence points, it
generates a simple model of occurence based on the distribution of those
points (i.e., the likelihood of an occurence at a location is inversely
proportional to the distance of that location from known occurences).
geoDist returns a raster of ‘suitability’ values that can be evaluated
just as the output from an SDM model projection.
Projection
Complex models, with large numbers of predictors, or that use complex features, are more likely to be overfit. Overfitting sacrifices generality (i.e., capacity to project to different scenarios) in favour of better fit to training data. Consequently, for the purposes of projection, we may want to emphasize smaller, simpler models. See also Petitpierre et al. (2017) for discussion of variable selection.
Guisan et al. (2017): two related issues to consider when comparing the environment in the training region to that in the region into which the model is projected:
availability: are environment ‘types’ similarly abundant and available?
analog: are the environment ‘types’ in the projected range also present in the training range?
The act of projection implicitly assumes environments are fully analagous with equal availability.
This is to a large extent unavoidable, so we have to take measures to reduce it in data preparation, and/or account for it in interpretation of results.
Clamping
Maxent includes the option of “clamping” projections. This constrains the values for environmental values in the projected range to the limit of that variable that is found in the training range. This has the effect of setting the predicted value of all non-analog cells to the value for the most extreme environments that are found in the training region.
This will reduce the occurence of unrealistic patterns emerging from the extension of complex models beyond the range of values they were trained on. It’s probably better than not clamping, but no reason to expect it’s particularly realistic.
Guisan et al. (2017) doesn’t mention clamping at all. Instead, they recommend explicitly identifying non-analog environments and excluding them from the projection; or at least, identifying them clearly and giving them due consideration in the interpretation of the results.
MESS
Multivariate Environmental Similarity Surfaces, Elith et al. (2010), provide a
way to identify non-analog environments, and to quantify the extent to
which they differ from environments in the training area. The functions
dismo::mess and ecospat::ecospat.messpredicts::mess (as of
2023-10-19) is the main R function for calculating MESS values.
MESS is applied in geographic space, and calculates a single index value for each cell. The value is based on the single environmental variable that is most different from the conditions in the training region. If all variables at a cell are within the range of the training data, the MESS index will be between 0 and 1, with 1 indicating maximum similarity with the training range. Values below 0 indicate locations where at least one variable is outside the range of the training data. The further the departure from the training range, the lower the value will be. Note that this incorporates only the single worse variable: if only one variable is out of range, or all variables are out of range, the index only reflects the worst one.
There is no formal test or strict rule for interpreting MESS maps, other than that we ought to be skeptical of projected results into areas with MESS values < 0.
Extrapolation Detection
Extrapolation Detection, or exDet, was proposed by Mesgaran et al. (2014). It
uses the Mahalanobis distance as the basis for an index of novelty. It
accomodates covariation among variables, a major improvement over MESS.
I’ve put together a tutorial for completing exDet analysis in
R with more details.
COUE
Broennimann et al. (2012) presents and approach to contrast environmental
conditions between training and projection regions in E-space. E-space and
G-space provide complementary views of the niche. E-space shows the
environmental distribution of a species in the context of all possible
environmental combinations; G-space shows the geographic distribution of
the species, constrained to environmental conditions that actually exist in
the landscape. This analysis applies the COUE framework (centroid,
overlap, underfilling and expansion).
ecospat (Cola et al., 2017) provides all the functions necessary to
implement these analyses.
Climate Models
Climate projections are not predictions of future conditions—they are model-derived descriptions of possible future climates under a given set of plausible scenarios of climate forcings. The intention of simulating future climate is not to make accurate predictions regarding the future state of the climate system at any given point in time but to represent the range of plausible futures and establish the envelope that the future climate could conceivably occupy.
– Harris et al. (2014)
Recommendations
From Harris et al. (2014) (originally presented as 9 points):
Include a high and a low emissions scenario, to capture ‘best-case’ and ‘worst-case’ scenarios.
Note that as of 2014 we are trending close to the high emissions RCP8.5 scenario. RCP2.6 represents an increasingly unlikely aggressive mitigation approach.
Time and resources may limit us to one or two emissions scenarios, but more than one GCM should be used.
Different GCMs have different strengths, weaknesses and biases. Some models are known to be ‘wet’ or ‘dry’ or ‘hot’, compared to the mean of all GCMs. These biases are also spatially variable: some GCMs may be ‘hot’ for Africa, and ‘cold’ for North America.
Ostenibly different GCMs may share code and assumptions, and thus share biases in their projections.
Consider the most appropriate way to present the output from multiple climate models;
- If the multi-model mean is presented, report the range or standard deviation;
- best-case and worst-case scenarios can be presented using envelopes/binary maps: envelopes based on locations where any of the models predict suitable habitat show worst case (i.e., all areas identified as being suitable habitat in any model), envelopes based on locations identified by all models show best case (i.e., only areas that all models agree are suitable are identified) [good approach for IAS].
Choose a baseline time period appropriate to the study
- baselines are preferably defined on data amalgamated over 30 years, to account for stochastic inter-annual variation. Shorter time periods may be unacceptably influenced by noisy weather.
- baseline climate should correspond to the time period in which the data (i.e., observations) were collected
- NB: different climate sources use different baseline periods!
- Further discussed in Roubicek et al. (2010). They tested the sensitivity of SDMs trained on a different baseline than the one used to simulate the data, and found those models were signficantly worse than ones trained on the correct baseline.
Be aware of the real resolution of the climate data used;
- GCMs are coarse resolution, and need to be down-scaled for use with SDMs.
Maintain a dialog with climate modelers, to keep up-to-date with developments in climate models.
WorldClim
WorldClim 2.1 has CMIP6 data: 9 GCMs for four SSP, at resolutions down to 2.5 minutes (30 seconds overdue for release in March 2020), projected to 2040, 2060, 2080 and 2100.
WorldClim 1.4 has CMIP5 data: 19 GCMs, four RCP, projected to 2050 and 2070, at resolutions as low as 30 seconds.
Definitions
- AR
- Assessment Report for the IPCC. AR4 was released in 2007, AR5 in 2014, AR6 is scheduled for 2022
- CMIP
- Coupled Model Intercomparison Project, IPCC collection of GCMs, Numbered to correspond to the AR (eg. CMIP5 for AR5). “Models are only admitted to the CMIP archive if they meet a suite of rigorous requirements, including consistency with relevant observations, both past and present, and with fundamental physical principles.” IPCC doesn’t judge the models beyond being deemed fit for service. As such, the collection of models represents set of plausible future climates under a given emissions scenario.
- GCM
- General Circulation Model: 3D numerical representation of the climate system. 50-250km cell size, 10-20 layers in the atmosphere. Includes Atmosphere-Ocean GCM (AOGCM), incorportating interactions between the oceans and atmosphere, and Earth System Models (ESM), GCMs that incorporate biogeochemical cycles (eg. carbon cycle); ESM may also contain dynamic global vegetation models (DGVM)
- SRES / RCP / SSP
- Special Report on Emissions Scenarios, socioeconomic analysis of future climate emissions under varying conditions, used in AR4. Updated to Representative Concentration Pathways, in AR5. Updated to Shared Socionomic Pathways, in AR6. RCP2.6/ SSP 126 is aggressive mitigation (best case); RCP8.5/SSP 585 is our current trajectory (worst case)
Downscaling
GCMs have a native resolution in the range of 50-250km2, much coarser than what is used for SDMs. To use for model projections, they need to be downscaled (i.e., converted to finer resolution) to the required scale. If you are using WorldClim data, it has already been downscaled for you.
Three options:
- Dynamic Downscaling
- uses the coarse-resolution GCMs as the input for similarly complex fine-scale models for the region of interest. Lots of added value compared to the GCMs, but computationally expensive, requiring expert skill to produce and interpret. Thus availability is limited.
- Statistical Downscaling
- uses past coars and fine-scale data to establish statistical/numerical models mapping regional data to local data. Then uses the resulting model to create future local data from future projections. Easier to do than Dynamic Downscaling, but less realistic, more implicit assumptions.
- Simple Scaling
- break coarse pixels up into smaller pixels containing the same value as the parent; doesn’t create any new values, or introduce any error.
Ensembles
TODO
TO-READ
Boria, R. A. et al. 2014. Spatial filtering to reduce sampling bias can improve the performance of ecological niche models. – Ecol. Model. 275: 73–77.
References
Aiello‐Lammens, M. E., R. A. Boria, A. Radosavljevic, B. Vilela, and R. P. Anderson. 2015. spThin: an R package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 38: 541–545.
Atwater, D. Z., C. Ervine, and J. N. Barney. 2018. Climatic niche shifts are common in introduced plants. Nature Ecology & Evolution 2: 34–43.
Bahn, V., and B. J. McGill. 2007. Can niche-based distribution models outperform spatial interpolation? Global Ecology and Biogeography 16: 733–742.
Barbet-Massin, M., F. Jiguet, C. H. Albert, and W. Thuiller. 2012. Selecting pseudo-absences for species distribution models: how, where and how many? Methods in Ecology and Evolution 3: 327–338.
Barve, N., V. Barve, A. Jiménez-Valverde, A. Lira-Noriega, S. P. Maher, A. T. Peterson, J. Soberón, and F. Villalobos. 2011. The crucial role of the accessible area in ecological niche modeling and species distribution modeling. Ecological Modelling 222: 1810–1819.
Booth, T. H. 2022. Checking bioclimatic variables that combine temperature and precipitation data before their use in species distribution models. Austral Ecology 47: 1506–1514.
Boyce, M. S., P. R. Vernier, S. E. Nielsen, and F. K. A. Schmiegelow. 2002. Evaluating resource selection functions. Ecological Modelling 157: 281–300.
Broennimann, O., M. C. Fitzpatrick, P. B. Pearman, B. Petitpierre, L. Pellissier, N. G. Yoccoz, W. Thuiller, et al. 2012. Measuring ecological niche overlap from occurrence and spatial environmental data. Global Ecology and Biogeography 21: 481–497.
Cola, V. D., O. Broennimann, B. Petitpierre, F. T. Breiner, M. D’Amen, C. Randin, R. Engler, et al. 2017. ecospat: an R package to support spatial analyses and modeling of species niches and distributions. Ecography 40: 774–787.
Elith, J., C. H. Graham, R. P. Anderson, M. Dudík, S. Ferrier, A. Guisan, R. J. Hijmans, et al. 2006. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29: 129–151.
Elith, J., M. Kearney, and S. Phillips. 2010. The art of modelling range-shifting species. Methods in Ecology and Evolution 1: 330–342.
Elith, J., S. J. Phillips, T. Hastie, M. Dudík, Y. E. Chee, and C. J. Yates. 2011. A statistical explanation of MaxEnt for ecologists: Statistical explanation of MaxEnt. Diversity and Distributions 17: 43–57.
Fick, S. E., and R. J. Hijmans. 2017. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology 37: 4302–4315.
Guisan, A., W. Thuiller, and N. E. Zimmermann. 2017. Habitat Suitability and Distribution Models: with Applications in R. Cambridge University Press.
Harris, I., T. J. Osborn, P. Jones, and D. Lister. 2020. Version 4 of the CRU TS monthly high-resolution gridded multivariate climate dataset. Scientific Data 7: 109.
Harris, R. M. B., M. R. Grose, G. Lee, N. L. Bindoff, L. L. Porfirio, and P. Fox‐Hughes. 2014. Climate projections for ecologists. WIREs Climate Change 5: 621–637.
Hijmans, R. J., S. Phillips, J. Leathwick, and J. Elith. 2017. dismo R package Version 1.1-4.
Hirzel, A. H., G. Le Lay, V. Helfer, C. Randin, and A. Guisan. 2006. Evaluating the ability of habitat suitability models to predict species presences. Ecological Modelling 199: 142–152.
Lee‐Yaw, J. A., M. Fracassetti, and Y. Willi. 2018. Environmental marginality and geographic range limits: a case study with Arabidopsis lyrata ssp. lyrata. Ecography 41: 622–634.
Legendre, P., and L. Legendre. 2012. Numerical ecology. 3rd Edition. Elsevier, New York.
Lobo, J. M., A. Jiménez‐Valverde, and R. Real. 2008. AUC: a misleading measure of the performance of predictive distribution models. Global Ecology and Biogeography 17: 145–151.
Merow, C., M. J. Smith, and J. A. Silander. 2013. A practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography 36: 1058–1069.
Mesgaran, M. B., R. D. Cousens, and B. L. Webber. 2014. Here be dragons: a tool for quantifying novelty due to covariate range and correlation change when projecting species distribution models J. Franklin [ed.], Diversity and Distributions 20: 1147–1159.
Muscarella, R., P. J. Galante, M. Soley‐Guardia, R. A. Boria, J. M. Kass, M. Uriarte, and R. P. Anderson. 2014. ENMeval: An R package for conducting spatially independent evaluations and estimating optimal model complexity for Maxent ecological niche models. Methods in Ecology and Evolution 5: 1198–1205.
Pearson, R. G., S. J. Phillips, M. M. Loranty, P. S. A. Beck, T. Damoulas, S. J. Knight, and S. J. Goetz. 2013. Shifts in Arctic vegetation and associated feedbacks under climate change. Nature Climate Change 3: 673–677.
Petitpierre, B., O. Broennimann, C. Kueffer, C. Daehler, and A. Guisan. 2017. Selecting predictors to maximize the transferability of species distribution models: lessons from cross-continental plant invasions. Global Ecology and Biogeography 26: 275–287.
Phillips, S. J. 2017. A Brief Tutorial on Maxent. Website http://biodiversityinformatics.amnh.org/open_source/maxent/ [accessed 19 May 2020].
Phillips, S. J., R. P. Anderson, M. Dudík, R. E. Schapire, and M. E. Blair. 2017. Opening the black box: an open-source release of Maxent. Ecography 40: 887–893.
Phillips, S. J., and M. Dudík. 2008. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31: 161–175.
Phillips, S. J., and J. Elith. 2010. POC plots: calibrating species distribution models with presence-only data. Ecology 91: 2476–2484.
Radosavljevic, A., and R. P. Anderson. 2014. Making better Maxent models of species distributions: complexity, overfitting and evaluation. Journal of Biogeography 41: 629–643.
Rodríguez-Rey, M., A. Jiménez-Valverde, and P. Acevedo. 2013. Species distribution models predict range expansion better than chance but not better than a simple dispersal model. Ecological Modelling 256: 1–5.
Roubicek, A. J., J. VanDerWal, L. J. Beaumont, A. J. Pitman, P. Wilson, and L. Hughes. 2010. Does the choice of climate baseline matter in ecological niche modelling? Ecological Modelling 221: 2280–2286.
Soberón, J. M. 2010. Niche and area of distribution modeling: a population ecology perspective. Ecography 33: 159–167.
Valavi, R., G. Guillera-Arroita, J. J. Lahoz-Monfort, and J. Elith. 2022. Predictive performance of presence-only species distribution models: a benchmark study with reproducible code. Ecological Monographs 92: e01486.
Varela, S., R. P. Anderson, R. García-Valdés, and F. Fernández-González. 2014. Environmental filters reduce the effects of sampling bias and improve predictions of ecological niche models. Ecography 37: 1084–1091.
Warren, D. L., N. Matzke, M. Cardillo, J. Baumgartner, L. Beaumont, N. Huron, M. Simões, et al. 2019. ENMTools R Package.
Warren, D. L., and S. N. Seifert. 2011. Ecological niche modeling in Maxent: the importance of model complexity and the performance of model selection criteria. Ecological Applications 21: 335–342.